|
| | Molecular_rExp_tExp () |
| |
| | Molecular_rExp_tExp (const Molecular_rExp_tExp &modello) |
| |
| void | stationary_dyn (double dt, std::vector< double > &m0, int changed_pos) |
| |
| void | change_par (int change) |
| |
| void | change_molecular () |
| |
| int | agg_dyn (double dt, double t) |
| |
| void | agg_dyint (double dt, double t) |
| |
| void | save_dyn (std::ofstream &file_, double t) |
| |
| void | gnuplot_single (string *names_files_Ecoli_mod, string &names_indice_mod, string *names_files_tau_mod, string &names_file_dyn_mod, string *names_info_mod, double T_f, Funz_C *f, int save_, int con_gen_sim) |
| |
| void | print_info (std::ofstream &file_save) |
| |
| void | reset_par () |
| |
| | ParameterEstimation_E_coli () |
| |
| | ParameterEstimation_E_coli (const E_coli &modello) |
| |
| void | s_F_lambda_r (vector< Q_tau_stat > vector_lambda, int n_vect) |
| |
| void | set_GoodnessFit (int Good, double D_n, double cumD_n) |
| |
| | E_coli () |
| |
| | E_coli (const E_coli &modello) |
| |
| int | initial_position_ec (int j, std::array< double, 2 > x0, double Raggio, int num_dist, int &cont_dist_5, int delta_dist_cont, double Delta_delta_dist) |
| |
| int | dist_iniz_ec (std::array< double, 2 > x0, double R, unsigned int num_dist) |
| |
| std::array< double, 2 > | X () |
| |
| double | V () |
| |
| int | N_dyn_var () |
| |
| double | C () |
| |
| double | C_iniziale () |
| |
| double | Production_rate () |
| |
| void | setProductionRate (double _prodRate) |
| |
| void | Tau_t (double tt_) |
| |
| void | Get_tipo_response_c (int T_r) |
| |
| void | Get_RC_0 (double Rc0) |
| |
| void | Get_RC_1 (double Rc1) |
| |
| void | Get_RC_q (int Rcq) |
| |
| int | F_tipo_response_c () |
| |
| double | F_RC_0 () |
| |
| double | F_RC_1 () |
| |
| double | F_RC_q () |
| |
| void | set_response (int ftipo_response_c, double fRC_0, double fRC_1, int fRC_q) |
| |
| void | agg_ligand (double t, Funz_C *f) |
| |
| void | aggiornamento (double dt, double t, Funz_C *f, int &sign_p, ofstream *file_tau, std::ofstream &file_theta) |
| |
| void | start_simulation (Funz_C *f) |
| |
| void | response_function () |
| |
| void | save_E_coli (ofstream *file, double t) |
| |
| void | save_E_coli_initial (std::ofstream *file_, double t) |
| |
| void | save_run (double t, ofstream &file_salto, double dt) |
| |
| void | save_tumble (double t, ofstream &file_tumble, double dt) |
| |
| void | save_theta (double t, double theta, std::ofstream &file_run) |
| |
| virtual int | saveIntDyn (std::string nameFile) |
| |
| virtual void | produce (Funz_C *f_i, double dt) |
| |
| virtual double | reset_barrier () |
| |
| virtual double | reset_barrier_t () |
| |
| virtual void | save_dyn (ofstream &file, double t) |
| |
| virtual void | gnuplot_single (std::string *names_files_Ecoli_mod, std::string &names_indice_mod, std::string *names_files_tau_mod, std::string &names_file_dyn_mod, std::string *names_info_mod, double T_f, Funz_C *f, int save_, int con_gen_sim) |
| |
| virtual void | gnuplot_single_film (std::string *names_files_Ecoli_mod, std::string &names_indice_mod, std::string *names_files_tau_mod, std::string &names_file_dyn_mod, std::string *names_info_mod, double T_f, Funz_C *f, int cont_sim) |
| |
| virtual void | gnuplot_single_film_gif (std::string *names_files_Ecoli_mod, std::string &names_indice_mod, std::string *names_files_tau_mod, std::string &names_file_dyn_mod, std::string *names_info_mod, double T_f, Funz_C *f, int cont_sim) |
| | produce the .gif film More...
|
| |
| virtual void | gnuplotFunzInternalDynalmic (std::string buffer, int save_, int con_gen_sim) |
| |
| virtual void | gnuplot_response (std::string names_info_mod, int save_, int con_gen_sim) |
| |
| virtual void | s_F_lambda_r (std::vector< Q_tau_stat > vector_lambda, int n_vect) |
| |
| void | statF_lambda_r (s_lambda &vector_lambda) |
| |
| virtual void | debugFunction () |
| |
|
| int | writeScriptgnuplotSingle (string tipo, string *names_files_Ecoli_mod, string *names_files_tau_mod, string *names_info_mod, string &names_file_dyn_mod, double T_f, int con_gen_sim, int save_) |
| |
| void | setSeed_ec (unsigned int *seed_thread) |
| |
| virtual int | writeScriptgnuplotSingle (std::string tipo, std::string *names_files_Ecoli_mod, std::string *names_files_tau_mod, std::string *names_info_mod, std::string &names_file_dyn_mod, double T_f, int con_gen_sim, int save_) |
| |
| virtual int | writeScriptLunchgnuplotSingle (std::string tipo, int save_, int con_gen_sim) |
| |
| double | Exp_dist_ec () |
| |
| double | rand_normal_ec (double stddev) |
| |
| double | gamma_par_double_ec () |
| |
| double | unifRand_ec () |
| |
| double | deltaW_ec (double dt) |
| |
| double | newtheta_ec (double theta) |
| |
| void | setEngine_ec (std::mt19937_64 *engine_thread_theta, std::mt19937_64 *engine_thread_bar, std::mt19937_64 *engine_thread_altro) |
| |
Celani Shimitzu Vergassola Molecular Model Class for a bacterium.
Class implementing Celani Shimitzu Vergassola Molecular model
This model is taken from CSV11 [A. Celani, T.S. Shimizu and M. Vergassola, Molecular and functional aspects of bacterial chemotaxis, J. Stat. Phys. 144, 219-240 (2011).], and implement the mean field approximation for the molecular-base model of the internal dynamic of the bacterium. The generalization to the Inverse gaussian and exponential-inverse gaussian is straightforward.
The chemotactic transduction pathway is conveniently modeled by the following set of mean-field equations:
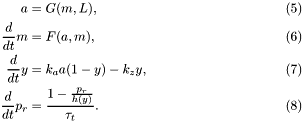
The detailed expressions for the functions that appear above are
\frac{M-a}{M-m+K_R}-k_b[\textrm{CheB}]a\frac{m}{m+K_B};\\ h(y)&=\frac{1}{1+\Big(\frac{y}{y_0}\Big)^H}. \end{align*}](form_16.png)


 Public Member Functions inherited from ParameterEstimation_E_coli
Public Member Functions inherited from ParameterEstimation_E_coli Public Member Functions inherited from E_coli
Public Member Functions inherited from E_coli Protected Member Functions inherited from E_coli
Protected Member Functions inherited from E_coli Protected Attributes inherited from ParameterEstimation_E_coli
Protected Attributes inherited from ParameterEstimation_E_coli Protected Attributes inherited from E_coli
Protected Attributes inherited from E_coli Public Attributes inherited from E_coli
Public Attributes inherited from E_coli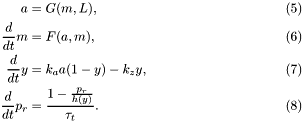
\frac{M-a}{M-m+K_R}-k_b[\textrm{CheB}]a\frac{m}{m+K_B};\\ h(y)&=\frac{1}{1+\Big(\frac{y}{y_0}\Big)^H}. \end{align*}](form_16.png)
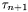 of the inernal variable lambda_[...] Heuristically we have:
of the inernal variable lambda_[...] Heuristically we have: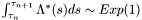
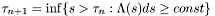
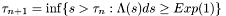
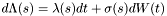 .
.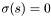 in the first case above.
in the first case above.