 |
EcoliSimulator
2.0.1a
|
 |
EcoliSimulator
2.0.1a
|
Base class for the function describing the ligand concentration. More...
#include <Funz_C.h>

Public Member Functions | |
| Funz_C () | |
| Funz_C (const Funz_C &f) | |
| virtual double | new_F_C (double t, std::array< double, 2 > x) |
| void | reset_parameter () |
| virtual void | set_parameter () |
| virtual void | get_coordinate (std::array< double, 2 > x, int *n) |
| virtual void | get_coordinate1 (std::array< double, 2 > x, int *n) |
| virtual void | all_informations (std::ofstream &file_save) |
| virtual void | info_stat (std::ofstream &file_stat) |
| virtual void | read_info_stat (int fchange_par, std::ifstream &file_stat, std::streampos &posizion) |
| virtual void | print_fc (std::ofstream &file_save, double t) |
| virtual void | print_info (std::ofstream &file_save) |
| virtual void | preview_F_C () |
| void | gnuplotFunC (double max_fc, double dt, int n_frames) |
Public Attributes | |
| int | change_par |
| double | D_c |
| double | max_x |
| double | min_x |
| double | max_y |
| double | min_y |
| double | degradation_rate |
| int | n_x |
| int | n_y |
| double | dx |
| double | dy |
| int | interact |
| double ** | f_c |
| double ** | q_c |
Static Public Attributes | |
| static int | num_funz = 9 |
Friends | |
| double | Histogram_3D_FunzC (Funz_C *f, double dt, int n_frame_max) |
Base class for the function describing the ligand concentration.
NOTE: If you add new functions, you should add the same in Funz_C::print_info
case 0:
0- Type Function: Gaussian $$c(t,x):= C_10*exp(-x^2/C_00)*(R<=C_20) = 6*exp(-pow(R,2)/10)*(R<=100)$$"
case 1:
1- Type Function: smoothed Jump $$c(t,x):= C_01+C_11*(t>C_21) = 1+2*(t>15)$$
case 2:
2- Type Function: Dancing Sombrero $$c(t,x):= ... $$
break;
case 3:
3- Type Function: Jump up/down $$c(t,x):= 1+C_13*(t<=C_03)+C_23*(t>C_03)*(t<=C_03+1)*pow(t-C_03,2)+C_23*(t>C_03+1)$$ $$ = 1+2*(t<=6)+7*(t>6)*(t<=6+1)*pow(t-6,2)+7*(t>6+1)$$
case 4:
4- Type Function: -----—> Maybe we should change this function Cambiare la funzione !! $$c(t,x):= 1+C_14*(t<C_04)+(t>=C_14)*(t<C_24)+C_34*(t>C_24)$$
case 5:
5- Type Function: Half-plane-Linear $$c(t,x):= C_05*(1+max(C_15,x[0])/C_15) = 3*(1+max(R,x[0])/2)$$
break;
case 7:
7- Type Function: time-Exponential $$c(t,x):= C_07*exp(C_27*t) = 18.2*exp(0.2*t)$$
default:
Otherwise Type Function: $$c(t,x):= Const = 3$$
The are of the experiment is
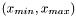
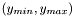
| Funz_C::Funz_C | ( | ) |
| Funz_C::Funz_C | ( | const Funz_C & | f | ) |
|
virtual |
save information for the report
|
virtual |
set in which the rectangle (dx_n,dy_n) of the mesh of the domain the point x fells
| n | is the array where the "coordinate" of the rectangle are saved |
|
virtual |
| void Funz_C::gnuplotFunC | ( | double | max_fc, |
| double | dt, | ||
| int | n_frames | ||
| ) |
draw function c
|
virtual |
save info for the statistical analysis
Reimplemented in Funz_C_Par.
|
virtual |
return the value of Ligand concentration in the point x
Reimplemented in Funz_C_Par, and Funz_C_Inter.
|
virtual |
previw of the dynamics of the ligand concentration (non-interacting case)
|
virtual |
save the mesh and relative value of Ligand in a file to draw with gnuplot
Reimplemented in Funz_C_Inter.
|
virtual |
similar to all_information
Reimplemented in Funz_C_Par, and Funz_C_Inter.
|
virtual |
read parameters from file
Reimplemented in Funz_C_Par.
| void Funz_C::reset_parameter | ( | ) |
reset parameters to the default
|
virtual |
set parameters of the ligand concentration in the Funz_C, you can choose the type of function and the dimantion of the rectangular area of the experiment
Reimplemented in Funz_C_Par, and Funz_C_Inter.
|
friend |
create hystogram for the preview
Function to create the histogram for the function c, it is used to PREVIEW LIGAND CONCENTRATION. (void Funz_C::preview_F_C())
| int Funz_C::change_par |
variable which track if the parameters has been changed: one might change int into boolean, 1 = change
| double Funz_C::D_c |
Diffusion coefficient: used in the interacting model
| double Funz_C::degradation_rate |
degradation rate of ligand see model in the population interacting with the ligand case
| double Funz_C::dx |
size-step mesh x direction
| double Funz_C::dy |
size-step mesh y direction
| double** Funz_C::f_c |
pointer to a matrix(n_x,n_y) with the value of the function: it is used to solve the PDE in the interacting case
| int Funz_C::interact |
== 0 se program interacting particles
| double Funz_C::max_x |
size of the area of the experiment
| double Funz_C::max_y |
| double Funz_C::min_x |
| double Funz_C::min_y |
| int Funz_C::n_x |
number od point in the mesh in the x direction (for interacting case)
| int Funz_C::n_y |
number od point in the mesh in the y direction (for interacting case)
|
static |
type of function c in the avaliable list
| double** Funz_C::q_c |
pointer to a matrix(n_x,n_y) with the value of the sorce: it is used to solve the PDE in the interacting case