 |
EcoliSimulator
2.0.1a
|
 |
EcoliSimulator
2.0.1a
|
Function used in the population interacting with the ligand setting. More...
#include <Funz_C_Inter.h>

Public Member Functions | |
| Funz_C_Inter () | |
| void | set_parameter () |
| double | new_F_C (double t, std::array< double, 2 > x) |
| void | print_fc (std::ofstream &file_save, double t) |
| void | print_info (std::ofstream &file_save) |
 Public Member Functions inherited from Funz_C_Par Public Member Functions inherited from Funz_C_Par | |
| Funz_C_Par () | |
| Funz_C_Par (const Funz_C_Par &f) | |
| double | new_F_C (double t, std::array< double, 2 > x) |
| void | set_parameter () |
| void | print_info (std::ofstream &file_save) |
| void | info_stat (std::ofstream &file_stat) |
| void | read_info_stat (int fchange_par, std::ifstream &file_stat, std::streampos &posizion) |
 Public Member Functions inherited from Funz_C Public Member Functions inherited from Funz_C | |
| Funz_C () | |
| Funz_C (const Funz_C &f) | |
| void | reset_parameter () |
| virtual void | get_coordinate (std::array< double, 2 > x, int *n) |
| virtual void | get_coordinate1 (std::array< double, 2 > x, int *n) |
| virtual void | all_informations (std::ofstream &file_save) |
| virtual void | preview_F_C () |
| void | gnuplotFunC (double max_fc, double dt, int n_frames) |
Additional Inherited Members | |
 Public Attributes inherited from Funz_C_Par Public Attributes inherited from Funz_C_Par | |
| double | C_0 |
| double | C_1 |
| double | C_2 |
| double | C_3 |
 Public Attributes inherited from Funz_C Public Attributes inherited from Funz_C | |
| int | change_par |
| double | D_c |
| double | max_x |
| double | min_x |
| double | max_y |
| double | min_y |
| double | degradation_rate |
| int | n_x |
| int | n_y |
| double | dx |
| double | dy |
| int | interact |
| double ** | f_c |
| double ** | q_c |
 Static Public Attributes inherited from Funz_C Static Public Attributes inherited from Funz_C | |
| static int | num_funz = 9 |
Function used in the population interacting with the ligand setting.
In case the program mode selected is population interacting with the ligand the Ligand concentration follows the following equation:
![\[ \frac{\partial}{\partial t}c(t,x,y)=D_c\Delta_{(x,y)}c(t,x,y)-K_c c(t,x,y)+K_p\sum_{i=1}^{n_c}\delta_{(t,x,y)}(p^i_b(t,x,y)) \]](form_19.png)
where 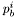 is the position of the bacterium number
is the position of the bacterium number 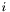 .
.
The simulation is performed using an ADI method where the boundary conditions are of Neumann's type. The method is as follows: Let 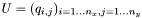 be the matrix of the mesh for the rectangular domain and
be the matrix of the mesh for the rectangular domain and  the matrix of the source. Let
the matrix of the source. Let 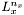 be the Laplacian-matrix is the
be the Laplacian-matrix is the 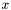 direction, it is a
direction, it is a 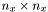 matrix (similar for
matrix (similar for 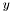 ), then using the Thompson's Algorithm for the resulting tridiagonal system we solve the PDE in two steps:
), then using the Thompson's Algorithm for the resulting tridiagonal system we solve the PDE in two steps:
![\[ L^{n_x}_x\otimes U = (U\otimes L^{n_y}_y)^T +\frac{dt\cdot Q}{2} \]](form_28.png)
![\[ L^{n_y}_y\otimes U^T = (U^T\otimes L^{n_x}_x)^T+\frac{dt\cdot Q^T}{2} \]](form_29.png)
where with 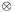 we indicate the
we indicate the matrix product, and with 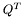 the transpose of
the transpose of 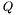 .
.
| Funz_C_Inter::Funz_C_Inter | ( | ) |
|
virtual |
it returns the value of the discretized version of c(t,x) on a grid
Reimplemented from Funz_C.
|
virtual |
save the mesh and relative value of Ligand in a file to draw with gnuplot
Reimplemented from Funz_C.
|
virtual |
similar to all_information
Reimplemented from Funz_C.
|
virtual |
set parameters of the ligand concentration in the Funz_C, you can choose the type of function and the dimantion of the rectangular area of the experiment
Reimplemented from Funz_C.