 |
EcoliSimulator
2.0.1a
|
 |
EcoliSimulator
2.0.1a
|
Base Class for a bacterium: It implements the virtual function to evolve the dynamic of the bacterium. More...
#include <E_Coli.h>
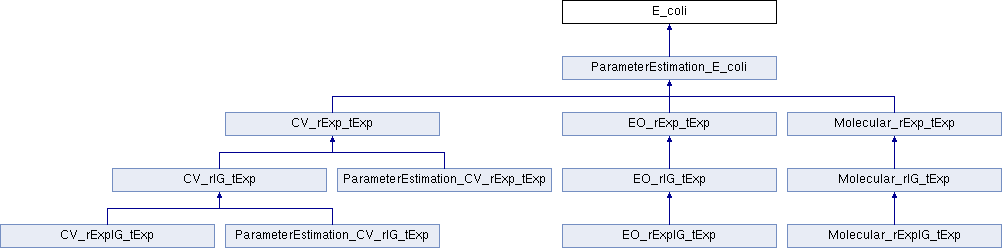
Public Member Functions | |
| E_coli () | |
| E_coli (const E_coli &modello) | |
| int | initial_position_ec (int j, std::array< double, 2 > x0, double Raggio, int num_dist, int &cont_dist_5, int delta_dist_cont, double Delta_delta_dist) |
| int | dist_iniz_ec (std::array< double, 2 > x0, double R, unsigned int num_dist) |
| std::array< double, 2 > | X () |
| double | V () |
| int | N_dyn_var () |
| double | C () |
| double | C_iniziale () |
| double | Production_rate () |
| void | setProductionRate (double _prodRate) |
| void | Tau_t (double tt_) |
| void | Get_tipo_response_c (int T_r) |
| void | Get_RC_0 (double Rc0) |
| void | Get_RC_1 (double Rc1) |
| void | Get_RC_q (int Rcq) |
| int | F_tipo_response_c () |
| double | F_RC_0 () |
| double | F_RC_1 () |
| double | F_RC_q () |
| void | set_response (int ftipo_response_c, double fRC_0, double fRC_1, int fRC_q) |
| void | agg_ligand (double t, Funz_C *f) |
| void | aggiornamento (double dt, double t, Funz_C *f, int &sign_p, ofstream *file_tau, std::ofstream &file_theta) |
| void | start_simulation (Funz_C *f) |
| void | response_function () |
| void | save_E_coli (ofstream *file, double t) |
| void | save_E_coli_initial (std::ofstream *file_, double t) |
| void | save_run (double t, ofstream &file_salto, double dt) |
| void | save_tumble (double t, ofstream &file_tumble, double dt) |
| void | save_theta (double t, double theta, std::ofstream &file_run) |
| virtual void | change_par (int change) |
| virtual void | reset_par () |
| virtual void | stationary_dyn (double dt, std::vector< double > &m0, int changed_pos) |
| virtual int | agg_dyn (double dt, double t) |
| virtual int | saveIntDyn (std::string nameFile) |
| virtual void | agg_dyint (double dt, double t) |
| virtual void | produce (Funz_C *f_i, double dt) |
| virtual double | reset_barrier () |
| virtual double | reset_barrier_t () |
| virtual void | save_dyn (ofstream &file, double t) |
| virtual void | gnuplot_single (std::string *names_files_Ecoli_mod, std::string &names_indice_mod, std::string *names_files_tau_mod, std::string &names_file_dyn_mod, std::string *names_info_mod, double T_f, Funz_C *f, int save_, int con_gen_sim) |
| virtual void | gnuplot_single_film (std::string *names_files_Ecoli_mod, std::string &names_indice_mod, std::string *names_files_tau_mod, std::string &names_file_dyn_mod, std::string *names_info_mod, double T_f, Funz_C *f, int cont_sim) |
| virtual void | gnuplot_single_film_gif (std::string *names_files_Ecoli_mod, std::string &names_indice_mod, std::string *names_files_tau_mod, std::string &names_file_dyn_mod, std::string *names_info_mod, double T_f, Funz_C *f, int cont_sim) |
| produce the .gif film More... | |
| virtual void | print_info (std::ofstream &file_save) |
| virtual void | gnuplotFunzInternalDynalmic (std::string buffer, int save_, int con_gen_sim) |
| virtual void | gnuplot_response (std::string names_info_mod, int save_, int con_gen_sim) |
| virtual void | set_GoodnessFit (int Good, double D_n, double cumD_n) |
| virtual void | s_F_lambda_r (std::vector< Q_tau_stat > vector_lambda, int n_vect) |
| void | statF_lambda_r (s_lambda &vector_lambda) |
| virtual void | debugFunction () |
Public Attributes | |
| std::mt19937_64 * | engine_theta |
| pointer to a engine (if I want to use std::uniform_real_distribution<double>) More... | |
| std::mt19937_64 * | engine_barrier |
| std::mt19937_64 * | engine_altro |
| int | simulation |
| std::string | Tipo_batterio |
| int | codice_batterio |
| s_lambda | s_lambda_r |
Protected Member Functions | |
| void | setSeed_ec (unsigned int *seed_thread) |
| virtual int | writeScriptgnuplotSingle (std::string tipo, std::string *names_files_Ecoli_mod, std::string *names_files_tau_mod, std::string *names_info_mod, std::string &names_file_dyn_mod, double T_f, int con_gen_sim, int save_) |
| virtual int | writeScriptLunchgnuplotSingle (std::string tipo, int save_, int con_gen_sim) |
| double | Exp_dist_ec () |
| double | rand_normal_ec (double stddev) |
| double | gamma_par_double_ec () |
| double | unifRand_ec () |
| double | deltaW_ec (double dt) |
| double | newtheta_ec (double theta) |
| void | setEngine_ec (std::mt19937_64 *engine_thread_theta, std::mt19937_64 *engine_thread_bar, std::mt19937_64 *engine_thread_altro) |
Protected Attributes | |
| double | production_rate |
| double | v |
| std::array< double, 2 > | x |
| double | theta |
| double | c |
| double | D_theta |
| double | lambda_r |
| double | lambda_t |
| double | tau_r |
| double | tau_t |
| int | n_dyn_var |
| double | barriera_r |
| double | barriera_t |
| int | salto_ |
| double | t_t |
| double | t_r |
| int | sign_c |
| int | up_down |
| double | c_iniziale |
| int | tipo_response_c |
| double | RC_0 |
| double | RC_1 |
| int | RC_q |
| int | tipoNewTheta |
| unsigned int * | seed_ecoli |
| pointer to a seed More... | |
Base Class for a bacterium: It implements the virtual function to evolve the dynamic of the bacterium.
| E_coli::E_coli | ( | ) |
| E_coli::E_coli | ( | const E_coli & | modello | ) |
|
virtual |
update internal dynamic
Reimplemented in CV_rExp_tExp, Molecular_rExp_tExp, and EO_rExp_tExp.
|
virtual |
update dynamic to control run/tumble
It controll the hitting time 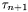 of the inernal variable lambda_[...] Heuristically we have:
of the inernal variable lambda_[...] Heuristically we have:
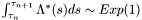
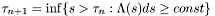
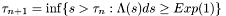
Where in general we can write: 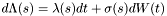 .
.
note 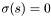 in the first case above.
in the first case above.
Reimplemented in CV_rExp_tExp, Molecular_rExp_tExp, EO_rExp_tExp, CV_rIG_tExp, EO_rIG_tExp, and Molecular_rIG_tExp.
| void E_coli::agg_ligand | ( | double | t, |
| Funz_C * | f | ||
| ) |
update ligand concentration
| void E_coli::aggiornamento | ( | double | dt, |
| double | t, | ||
| Funz_C * | f, | ||
| int & | sign_p, | ||
| ofstream * | file_tau, | ||
| std::ofstream & | file_theta | ||
| ) |
update all the parameters of the bacterium during the simulation
| double E_coli::C | ( | ) |
return current ligand concentration
| double E_coli::C_iniziale | ( | ) |
return initial ligand concentration
|
virtual |
set parameters model
Reimplemented in CV_rExp_tExp, Molecular_rExp_tExp, EO_rExp_tExp, CV_rIG_tExp, EO_rIG_tExp, and Molecular_rIG_tExp.
|
virtual |
Reimplemented in CV_rExp_tExp, and CV_rIG_tExp.
|
protected |
| int E_coli::dist_iniz_ec | ( | std::array< double, 2 > | x0, |
| double | R, | ||
| unsigned int | num_dist | ||
| ) |
|
protected |
| double E_coli::F_RC_0 | ( | ) |
return 1st coefficient
| double E_coli::F_RC_1 | ( | ) |
return 2nd coefficient
| double E_coli::F_RC_q | ( | ) |
return 3rd coefficient
| int E_coli::F_tipo_response_c | ( | ) |
return current type of response
|
protected |
| void E_coli::Get_RC_0 | ( | double | Rc0 | ) |
set 1st coefficient
| void E_coli::Get_RC_1 | ( | double | Rc1 | ) |
set 2nd coefficient
| void E_coli::Get_RC_q | ( | int | Rcq | ) |
set 3rd coefficient
| void E_coli::Get_tipo_response_c | ( | int | T_r | ) |
set type of response
| tt_ | type of response |
|
virtual |
plot response: empty function ... not yet implemented
Empty function
|
virtual |
plot via gnuplot trajectory and data for single tracking
Reimplemented in CV_rExp_tExp, and EO_rExp_tExp.
|
virtual |
save and produce film of single tracking
|
virtual |
produce the .gif film
save and produce film of single tracking in .gif
|
virtual |
plot internal dynamic
| int E_coli::initial_position_ec | ( | int | j, |
| std::array< double, 2 > | x0, | ||
| double | Raggio, | ||
| int | num_dist, | ||
| int & | cont_dist_5, | ||
| int | delta_dist_cont, | ||
| double | Delta_delta_dist | ||
| ) |
This function set the initial position. What it does is: if the distribution is the n. 5, then update the new center, otherwise it calls dist_iniz(x,x0,Raggio,num_dist).
| x[] | position of pacterium (to be set) |
| x0[] | initial position, set in the menu |
| R | radius distribution |
| type | of distribution, see menu |
| int E_coli::N_dyn_var | ( | ) |
return number of internal variables in the current model
|
protected |
|
virtual |
print info for LateX report
Reimplemented in CV_rExp_tExp, Molecular_rExp_tExp, EO_rExp_tExp, ParameterEstimation_CV_rExp_tExp, EO_rIG_tExp, CV_rIG_tExp, CV_rExpIG_tExp, and EO_rExpIG_tExp.
|
virtual |
update concentration ligand after production (in interacting mode)
| double E_coli::Production_rate | ( | ) |
retun rate of production of new ligand concentration by the bacterium
|
protected |
|
virtual |
reset trashold for the run
Reimplemented in EO_rIG_tExp, CV_rIG_tExp, Molecular_rIG_tExp, CV_rExpIG_tExp, Molecular_rExpIG_tExp, and EO_rExpIG_tExp.
|
virtual |
reset trashold for the tumble
|
virtual |
reset defolt parametes model
Reimplemented in CV_rExp_tExp, Molecular_rExp_tExp, EO_rExp_tExp, EO_rIG_tExp, CV_rIG_tExp, CV_rExpIG_tExp, Molecular_rExpIG_tExp, EO_rExpIG_tExp, and Molecular_rIG_tExp.
| void E_coli::response_function | ( | ) |
update the current concentration via the response function
|
virtual |
initialize and save the empirical satisfaction index (function lambda)
|
virtual |
save internal dynamic in a file
| void E_coli::save_E_coli | ( | ofstream * | file, |
| double | t | ||
| ) |
save position, concentration and direction to file
| void E_coli::save_E_coli_initial | ( | std::ofstream * | file_, |
| double | t | ||
| ) |
| void E_coli::save_run | ( | double | t, |
| ofstream & | file_salto, | ||
| double | dt | ||
| ) |
save time and duration of a run
| void E_coli::save_theta | ( | double | t, |
| double | theta, | ||
| std::ofstream & | file_run | ||
| ) |
function used to save the angle at the moment of a tumble.
| void E_coli::save_tumble | ( | double | t, |
| ofstream & | file_tumble, | ||
| double | dt | ||
| ) |
save time and duration of a tumble
| dt | time-step in the simulation |
|
virtual |
Reimplemented in CV_rExp_tExp, EO_rExp_tExp, EO_rIG_tExp, and CV_rIG_tExp.
|
virtual |
save outpot of goodness fit test
Reimplemented in ParameterEstimation_E_coli.
| void E_coli::set_response | ( | int | ftipo_response_c, |
| double | fRC_0, | ||
| double | fRC_1, | ||
| int | fRC_q | ||
| ) |
set response function
| type | and coefficient (from 1st to last) |
|
protected |
| void E_coli::setProductionRate | ( | double | _prodRate | ) |
set production rate (it is a static variable, once for all!)
|
protected |
| void E_coli::start_simulation | ( | Funz_C * | f | ) |
set initial data for the simulation
| void E_coli::statF_lambda_r | ( | s_lambda & | vector_lambda | ) |
|
virtual |
calculate stationary dynamics for the initial condition
| m0 | are the initial condition for the internal variables: if changed_pos says that the initial condition are not changed, for example if the initial distribution for the population is a delta dirac, then we just use the old values, and do not recalculate them |
Reimplemented in CV_rExp_tExp, Molecular_rExp_tExp, and EO_rExp_tExp.
| void E_coli::Tau_t | ( | double | tt_ | ) |
set time start of a tumble
| tt_ | time in sec. |
|
protected |
|
inline |
return speed bacterium
|
protectedvirtual |
write the script for the gnuplot of single bacterium
Reimplemented in CV_rExp_tExp, and EO_rExp_tExp.
|
protectedvirtual |
| array< double, 2 > E_coli::X | ( | ) |
return position bacterium
|
protected |
threshold for lambda_r (when lambda_r crosses it, tumble starts)
|
protected |
threshold for lambda_t (when lambda_r crosses it, run starts)
|
protected |
Ligand concentration
|
protected |
initial concentration of Ligand
| int E_coli::codice_batterio |
|
protected |
Diffusion coefficient direction
| std::mt19937_64* E_coli::engine_altro |
| std::mt19937_64* E_coli::engine_barrier |
| std::mt19937_64* E_coli::engine_theta |
pointer to a engine (if I want to use std::uniform_real_distribution<double>)
|
protected |
probability rate to tumble
|
protected |
probability rate to run
|
protected |
number internal variables
|
protected |
Production rate bacterium
|
protected |
1st paramter in the response function
|
protected |
2nd paramter in the response function
|
protected |
3rd paramter in the response function: esponent in th Hill function
| s_lambda E_coli::s_lambda_r |
estimated parameter for the function lambda
|
protected |
variable identifying in which state I'm in run_fase == 1, tumble_fase == 2, jump_instant == 0, tumble_fase == -1
|
protected |
pointer to a seed
The idea is the following: At the beginning of every simulation I seed the extra-thread
|
protected |
sign of the "temporal gradient": c(now) - c(now-dt)
| int E_coli::simulation |
MODE of simulation:
|
protected |
time began run
|
protected |
time began tumble
|
protected |
mean of a run in the steady state
|
protected |
mean of a tumble in the steady state
|
protected |
Direction
| std::string E_coli::Tipo_batterio |
Name of the model used
the format in as follows: [Name Model]_r[Name Distribution run]_t[Name Distribution tumble]
|
protected |
Type of reponse function between the available
|
protected |
This variable take track of the for of the distribution of the new theta
|
protected |
variable that keep track if I'm going up or down the gradient
|
protected |
Speed bacterium
|
protected |
Position